What Is DNA Sequencing?
The order of the four nucleotide bases (adenine, thymine, cytosine, and guanine) that make up the DNA molecule and carry essential genetic information is determined by a process called DNA sequencing. All four bases in DNA produce what are termed base pairs when they attach to their complementary partners (bp). The nucleotides adenine (A) and thymine (T) and cytosine (C) and guanine (G) couple with one another (G). About 3 billion base pairs make up the human genome and serve as the blueprint for building and maintaining a human being. DNA sequence’s base-paired nature makes it an excellent candidate for storing a large quantity of genetic information. Most techniques of DNA sequencing rely on this same complementary base-pairing that underpins the process by which DNA molecules are copied, transcribed, and translated. Complete genome sequencing was made practical and cheap by breakthroughs in DNA sequencing technology and methodologies.
Also read: alina orlova dog
DNA Sequencing Example
Once requiring years, DNA sequencing may now be completed in a matter of hours. More than $3 billion was spent to complete the first human DNA sequencing. The cost to get your whole genome sequenced by a company has dropped to below $1,000. The most cutting-edge diagnostic procedures will look at each every nucleotide in your DNA. Now however, numerous businesses provide SNP testing.
These analyses zero down on specific nucleotides inside genes that may represent a mutated form of a gene. These single-nucleotide polymorphisms (SNPs) are associated with specific diseases and may thus be used as potential biomarkers of an individual’s phenotype. Several single-nucleotide polymorphisms (SNPs) have been linked to specific illnesses, while others influence metabolic pathways and nutrition use. DNA sequencing may be used to learn how your genome impacts your life, and thousands of connections have already been discovered.
Different types of DNA sequencing technologies?
Some sequencing technologies have become obsolete since their inception in the 1970s (see the article what is DNA sequencing?), while others have advanced to become the standard for studying genetic diversity.
Sanger sequencing
When researchers need to do low-throughput, targeted, or short-read sequencing, they turn to Sanger sequencing. Sanger sequencing is still the method of choice for verifying variations found by orthogonal approaches and performing targeted sequencing due to its sensitivity and relative simplicity in terms of process and methodology.
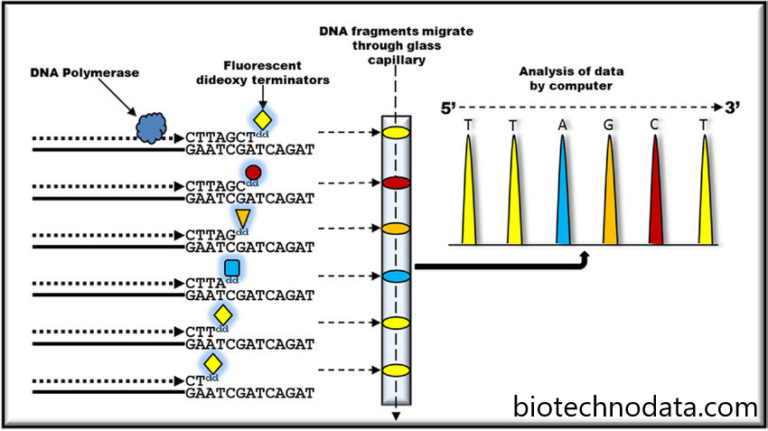
The Sanger sequencing technique employs a chain-termination strategy to reveal the specific sequence and identity of nucleotide bases along a DNA strand. Using chemical analogues of the four nucleotide bases, this technique is very accurate. Unlike their DNA counterparts, these ddNTPs lack the hydroxyl group necessary for polynucleotide chain expansion. Radiolabeled ddNTPs and template DNA are mixed together, and strands of varying lengths result from the random incorporation of the ddNTPs.
In the middle of the 1990s, scientists began employing capillary electrophoresis for Sanger sequencing. These technologies use long, thin, acrylic-fiber capillaries filled with a gel matrix to size-separate tagged DNA fragments. Electrokinetic injection of a sample containing the labelled fragments into the capillary is followed by the application of an electric field to elevate the fragments. The instrument’s internal detecting laser reads the labels as they pass it, allowing the sequence to be calculated as they go.
Capillary electrophoresis and fragment analysis
Sanger sequencing and fragment analysis are two applications that may be carried out using capillary electrophoresis (CE) devices. DNA fragments are fluorescently tagged, separated by CE, and measured against an internal standard in a process called fragment analysis. DNA sequencing by CE is typically used to determine a fragment’s or gene segment’s unique base sequence, but it can also be used to determine the size, relative quantitation, and genotyping of fragments of DNA that have been fluorescently labelled and generated via polymerase chain reaction with primers specific to the DNA target in question.
Authenticating cell lines and spotting aneuploidy are only two examples of the many uses made possible by analyzing DNA fragments. Researchers choose fragment analysis because of its shorter turnaround time, greater sensitivity and resolution, and lower cost compared to sequencing methods, which also allow for similar applications.
Next-generation sequencing (NGS)
New high-throughput methods, known as next-generation sequencing (NGS) technologies, have emerged alongside the many advancements made to Sanger sequencing over the years. The NGS process is quite parallel.
Depending on the objective, NGS may analyzes anywhere from a few of genes to a complete genome. The sequence of DNA bases across the whole genome or exome may be obtained using whole-genome sequencing (WGS) or whole-exome sequencing (WES). To evaluate changes and levels of gene expression across the transcriptome, whole-transcriptome sequencing offers sequence information on coding and numerous noncoding types of RNA. Only a select group of genes or areas of interest are sequenced during a focused sequencing project. Targeted sequencing is highly suited for use in both translational and clinical research because to its quick turnaround time, cheap cost, minimal sample input need, and relatively easy interpretation. It is common practice to employ Sanger sequencing to verify NGS-detected variations.
Also read: twitter ticketed spacespereztechcrunch
Applications of DNA Sequencing Technologies
The genetic information contained in a snippet of DNA, a whole genome, or a complicated microbiome may be deduced by sequencing the DNA that makes up these samples. Researchers may utilize sequence data to learn about the DNA molecule’s content, including the genes and regulatory instructions it contains. Gene-specific characteristics like open reading frames (ORFs) and CpG islands may be scanned for in the DNA sequence. Evolutionary studies between species or populations may be conducted by comparing homologous DNA sequences from various animals. In particular, DNA sequencing may identify variants in genes that are likely to be pathogenic.
DNA sequencing has been put to use in a variety of medical contexts, from illness diagnosis and therapy to epidemiological research. By enhancing agriculture via more precise plant and animal breeding and decreasing the dangers of disease outbreaks, sequencing has the potential to radically alter the state of food safety and sustainable agriculture. DNA sequencing also has applications in conservation and environmental improvement for the benefit of humans and other species.